在使用Python開發AI時,由於需時時查看處理中的訓練資料,於是大多使用Jupyter Notebook進行開發。這是利用Python是直譯式程式語言的的特點,在這裡就不說太多,到底要如何安裝Jupyter Notebook呢?
優點:免下載、已預先安裝大多數的函式庫、搞不好速度比你家電腦快、檔案會自動清除。
缺點:若想儲存資料須將資料下載或將Colab掛接到Google Drive、說不定比你家電腦慢、不能長時間閒置(會停止運算,但檔案還在)
點開連結:Colab
首次登入介面:
(未登入Google)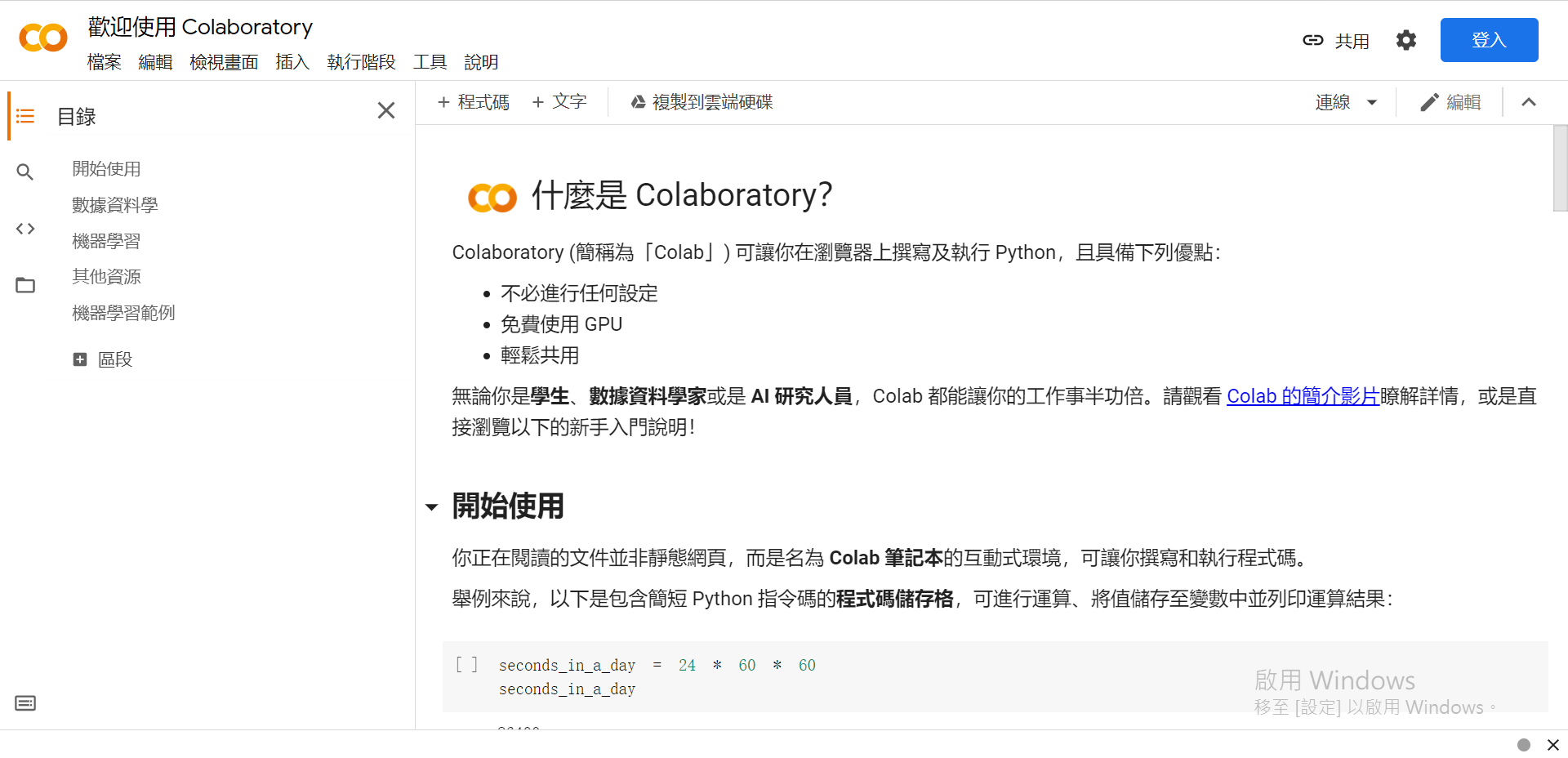
嘿對~我Windows還沒啟用,但我其實有買,這就是另一個故事了~
(已登入Google)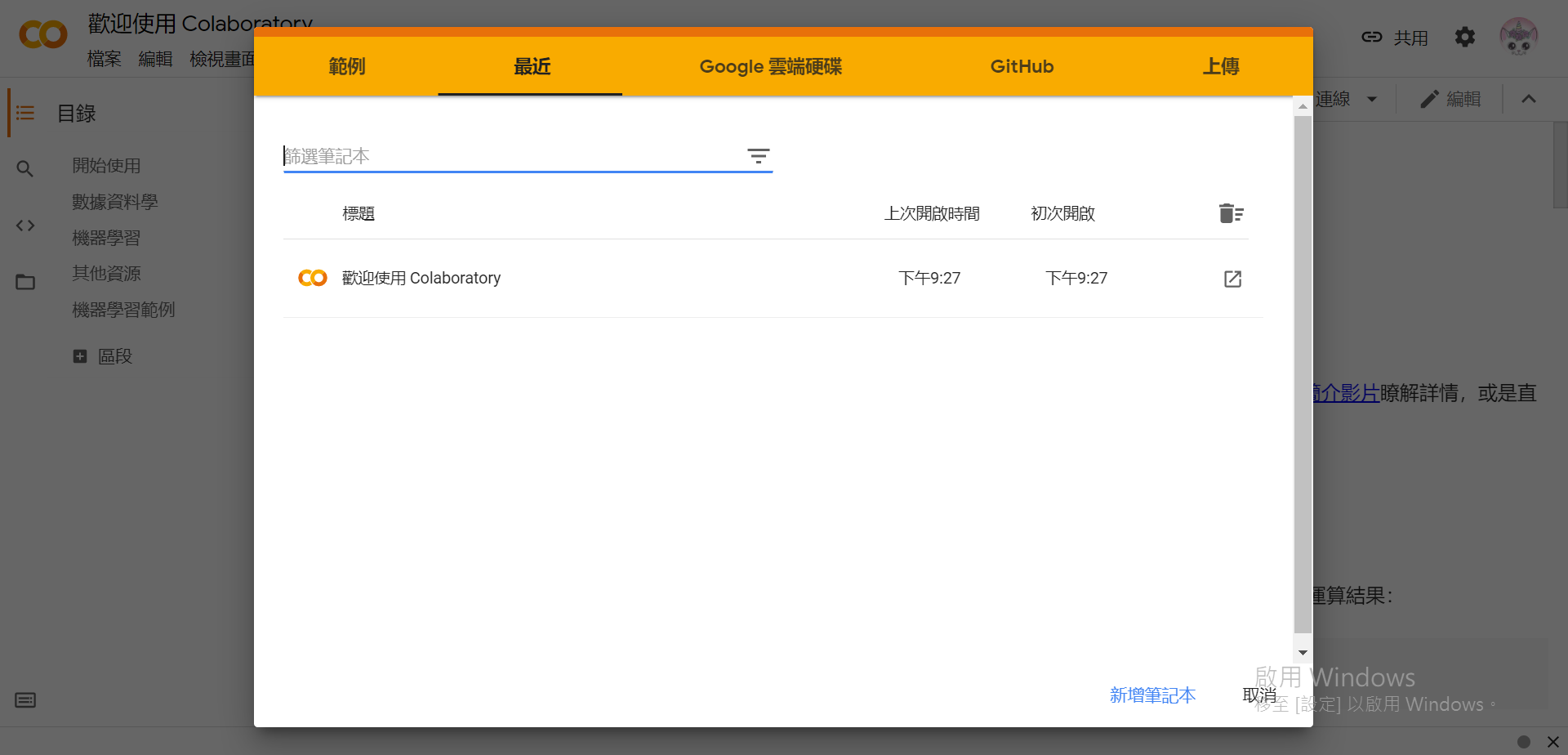
一共有5+1個選項:
.ipynb作為副檔名)。這裡我們選擇[取消],如果要儲存檔案的話可以之後在左上方的[檔案]中選擇儲存的位置。
如果覺得預設的範例很醜,可以點選第一個區塊(我們稱之為Cell),然後按垃圾桶進行刪除。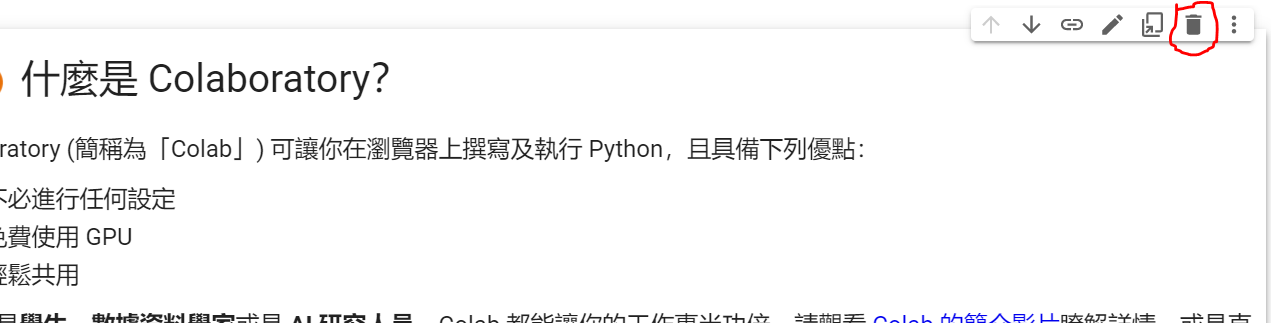
經一陣猛點後:
接著可以新增一個[+程式碼](Python)或[文字](MarkDown)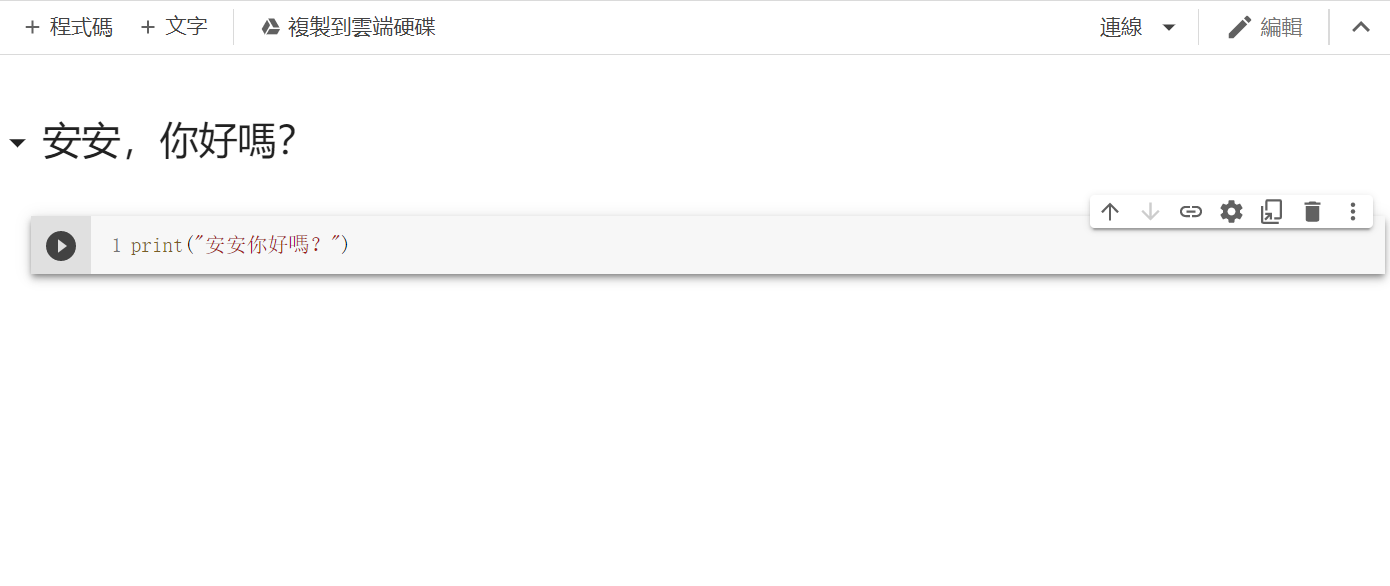
P.S:可以透過Cell右上方的箭頭來調整Cell的順序、
MarkDown的話可以點擊[關閉Markdown編輯器](在設定旁邊)來顯示內容。
再來點及右上方共用底下有一個連線,點他來連線到主機後,點選程式碼區塊左邊的執行儲存格。
結果: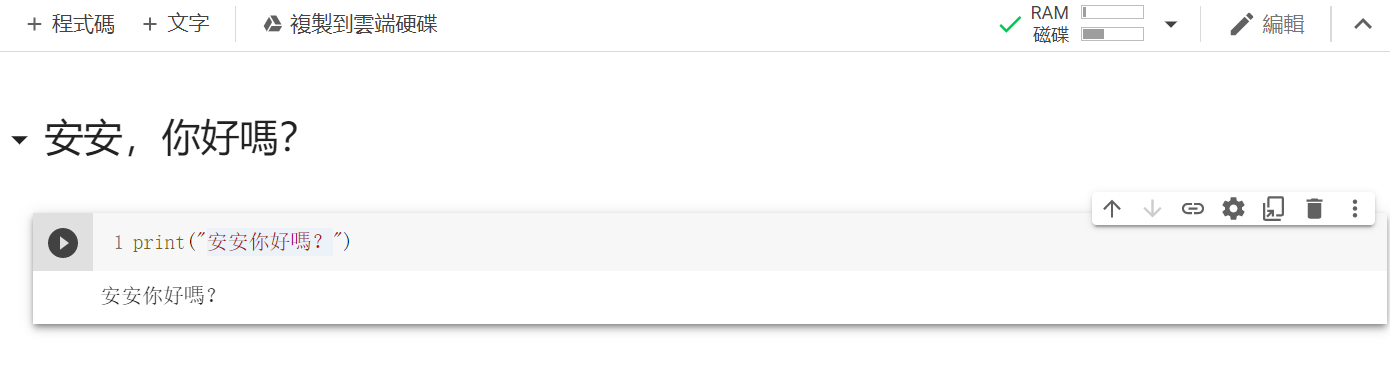
Google的Colab先到這邊,剩下的未來遇到再說,也可以先自行探索喔~(例如使用GPU加速、Google特有的TPU等等)
如果不想執行在雲端的話,另一個我推薦的是使用Anaconda IDE,他整合了許多資料科學套件,除了跑得有點慢,他會是你資料科學之旅的好朋友。
下載連結:Anaconda
能選64位元就選64位元,不然大量的數據灌下去你的電腦不知道要算到牛年馬月。(當然這取決於作業系統及硬體配置,詳細方法請見:如何查詢你的Windows系統是32或64位元?)
安裝完成後(就狂點下一步),初始畫面如下:
接著點選Environments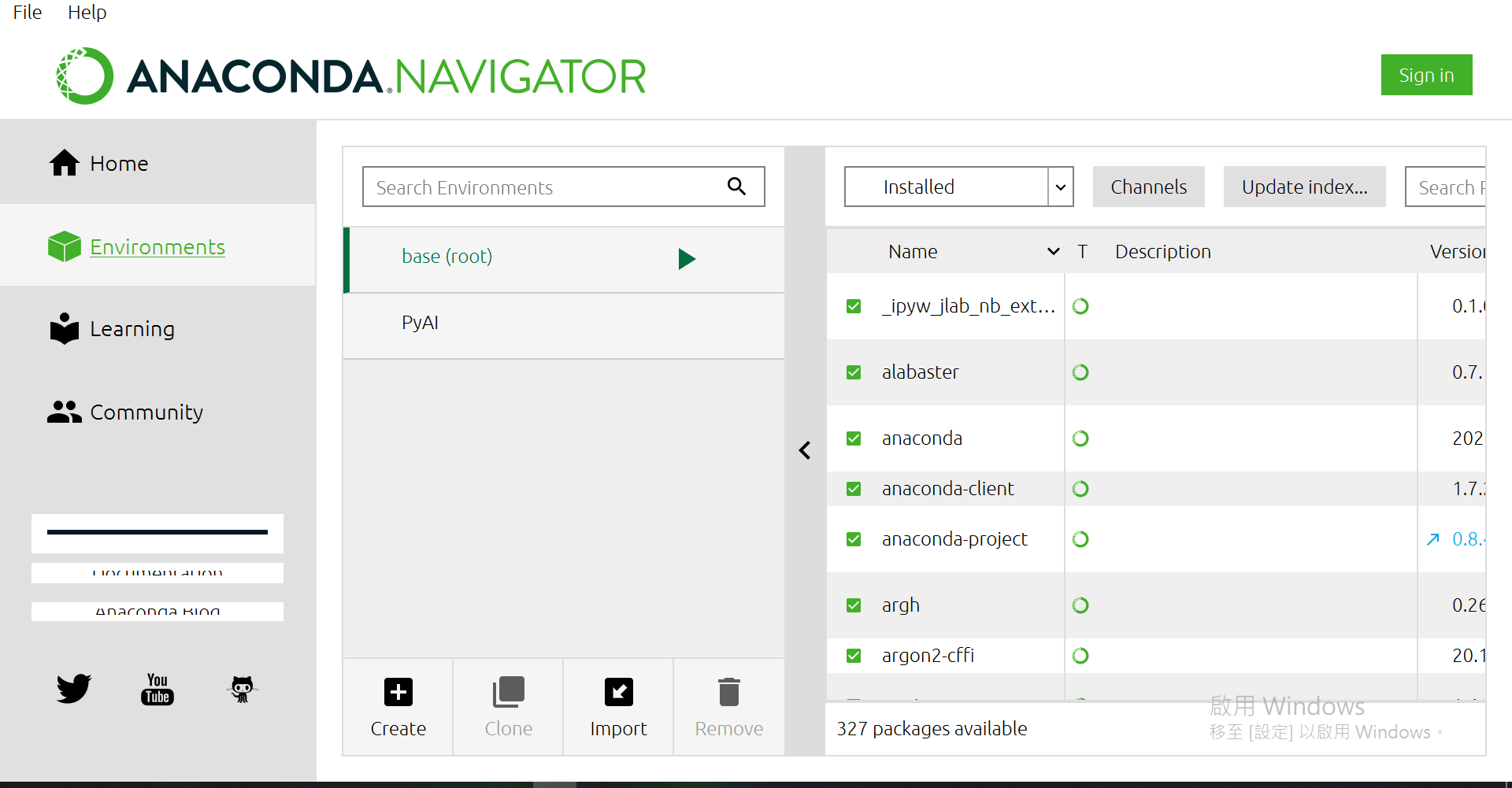
當然,你的畫面會沒有"PyAI"這個虛擬環境,現在就來創一個吧~
點選[Create]
打上自己想要的名子,版本我是選3.8啦~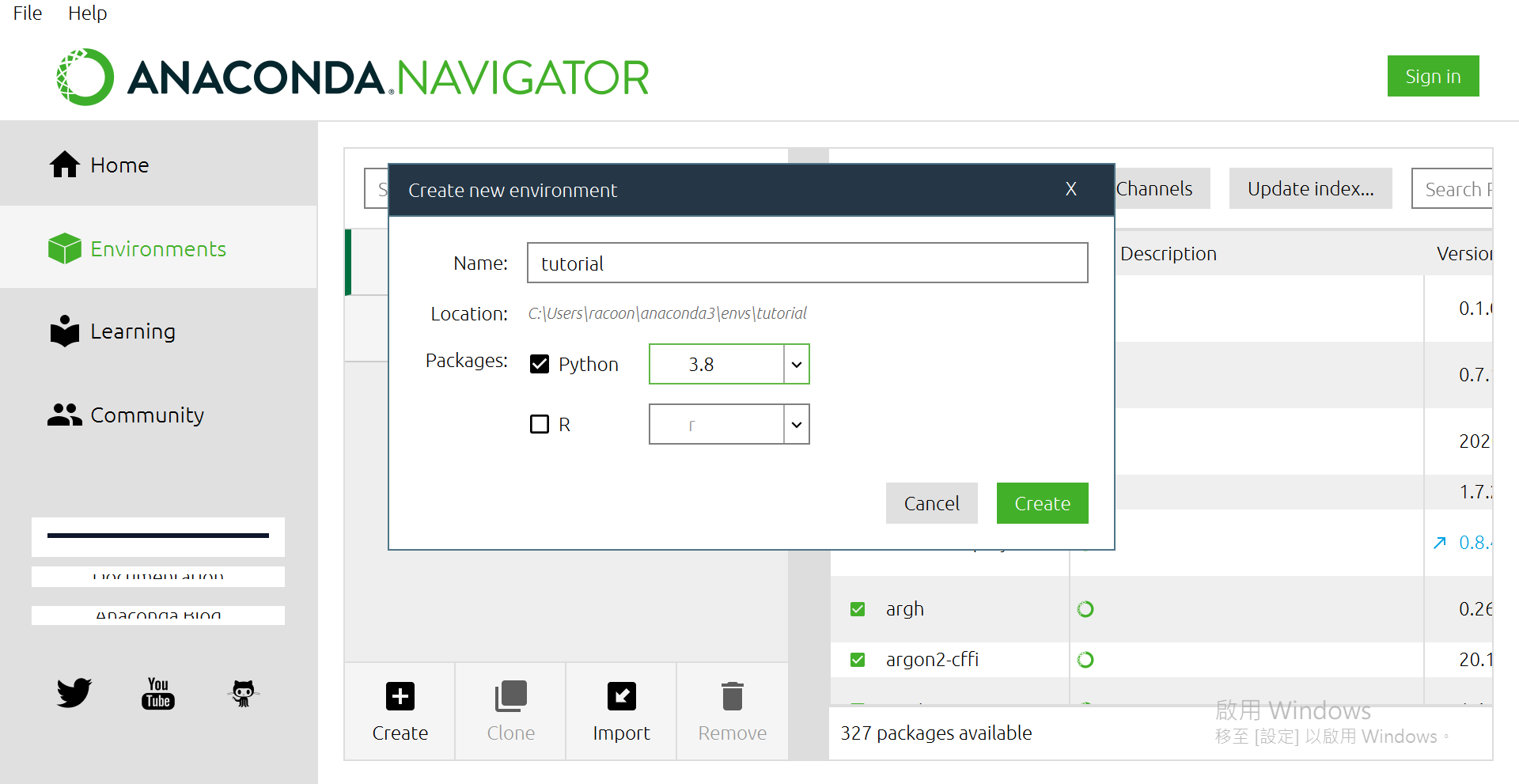
然後按[Create]
之後要選環境就可以進來這裡選,或是在首頁的地方有個"Applications on"可以切換。
接著安裝未來會用到的函式庫,保持在"Environments",中間有個箭頭"<"給他按下去。
有一個選單"Installed"選成"Not Installed",再來右邊可以搜尋,找到下面這幾個package給他點選起來:
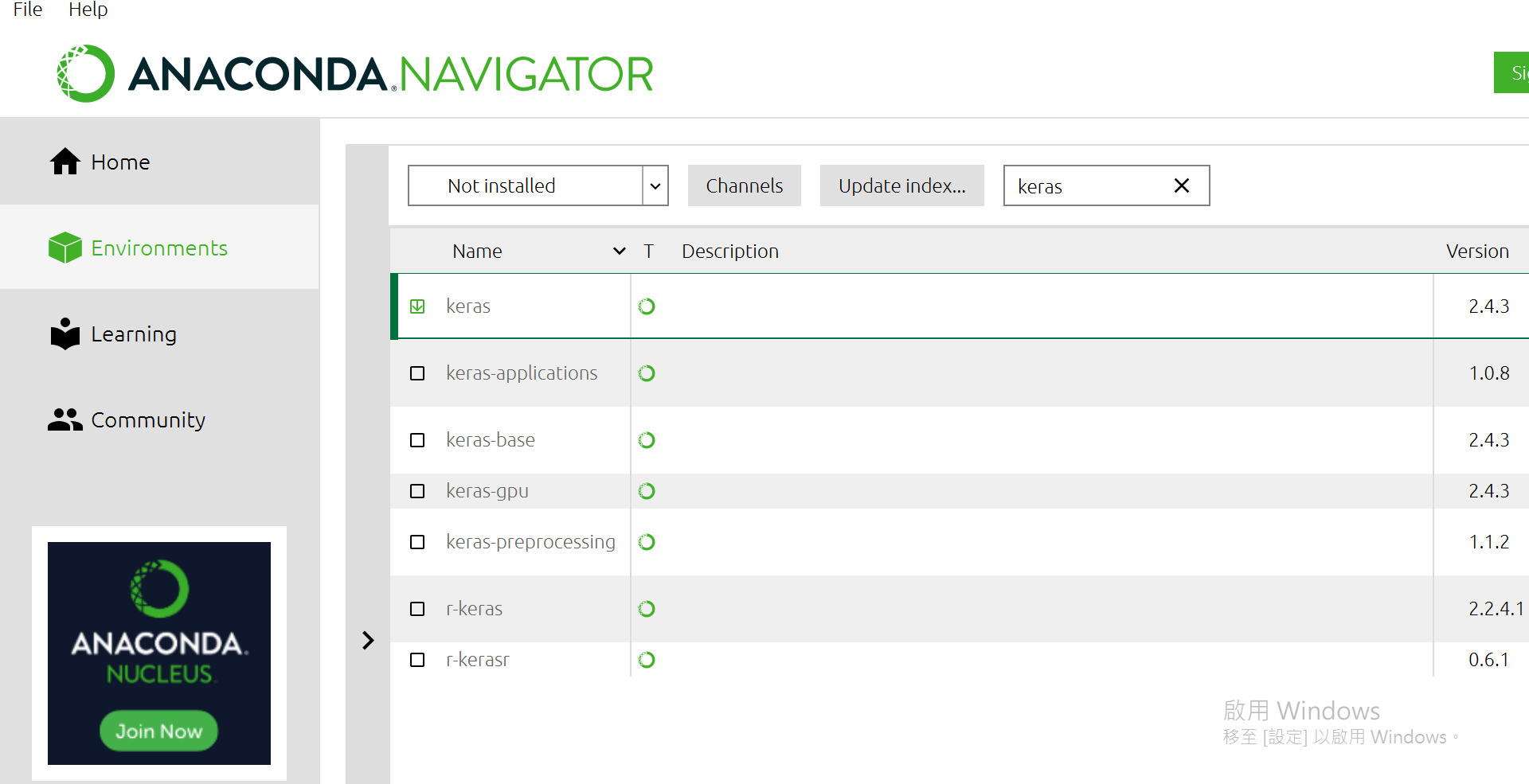
都選完後,右下方有個[Apply],按下去。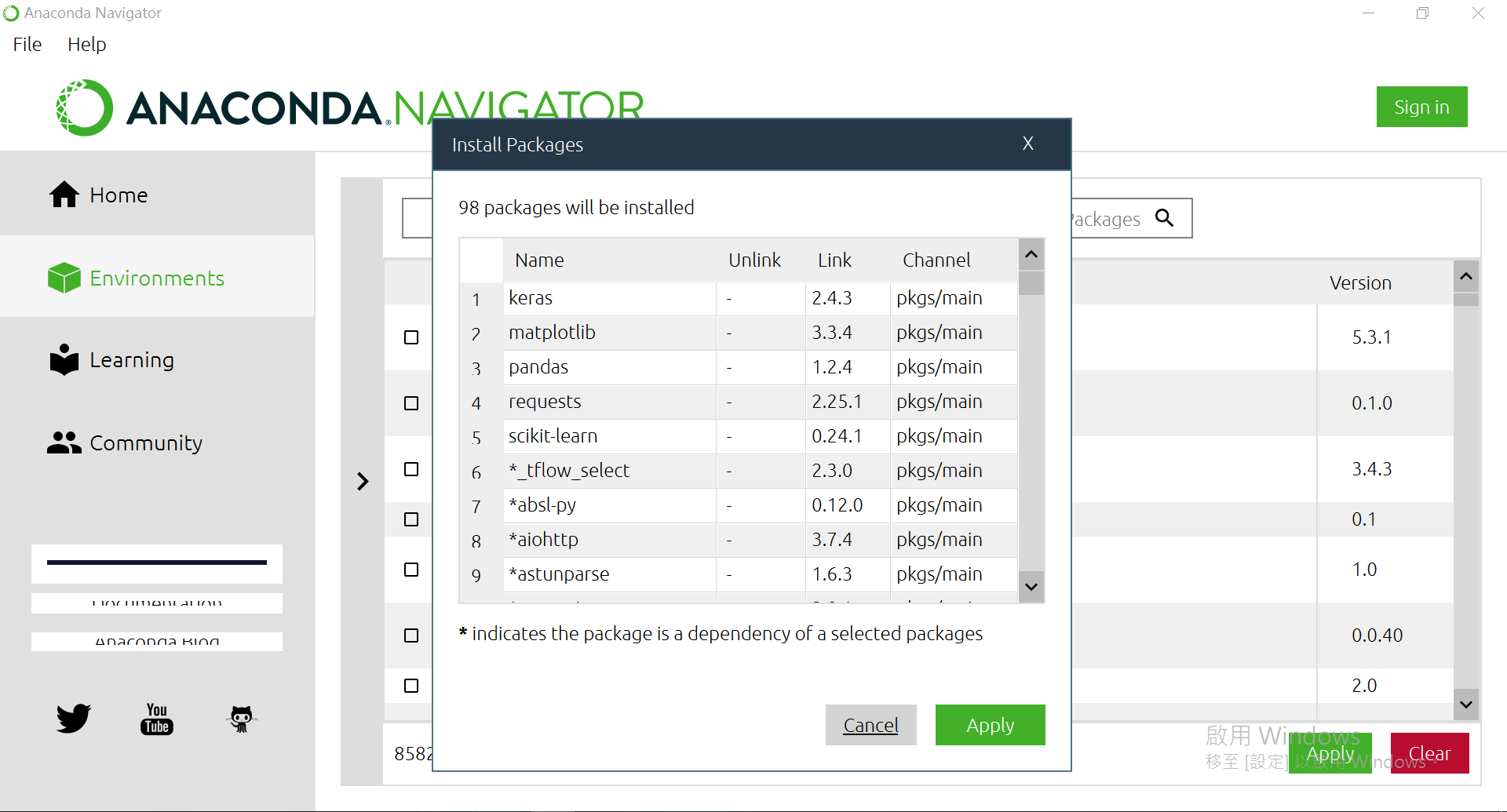
確認過眼神後,再[Apply]一次。
除了透過GUI的套件管理來下載函式庫外,也可以用終端機安裝。
把箭頭按回去後,點一下綠色三角形(記得要確認環境正確喔~不要到時候裝錯=.=)
點[Open Terminal]來使用pip進行安裝。
來安裝個套件看看: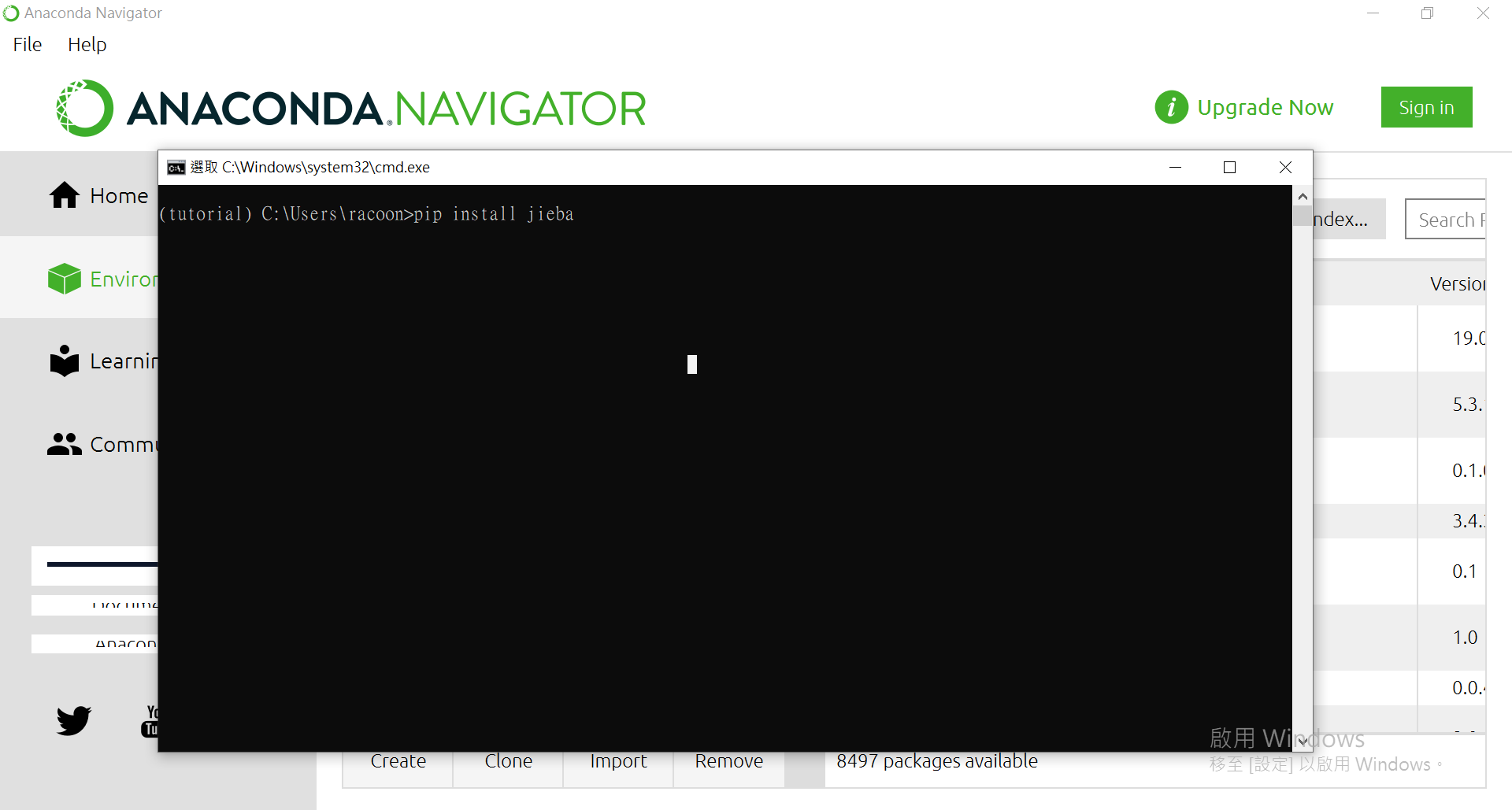
OK,回到主頁。往下滑找到Jupyter Notebook,並按下[Install]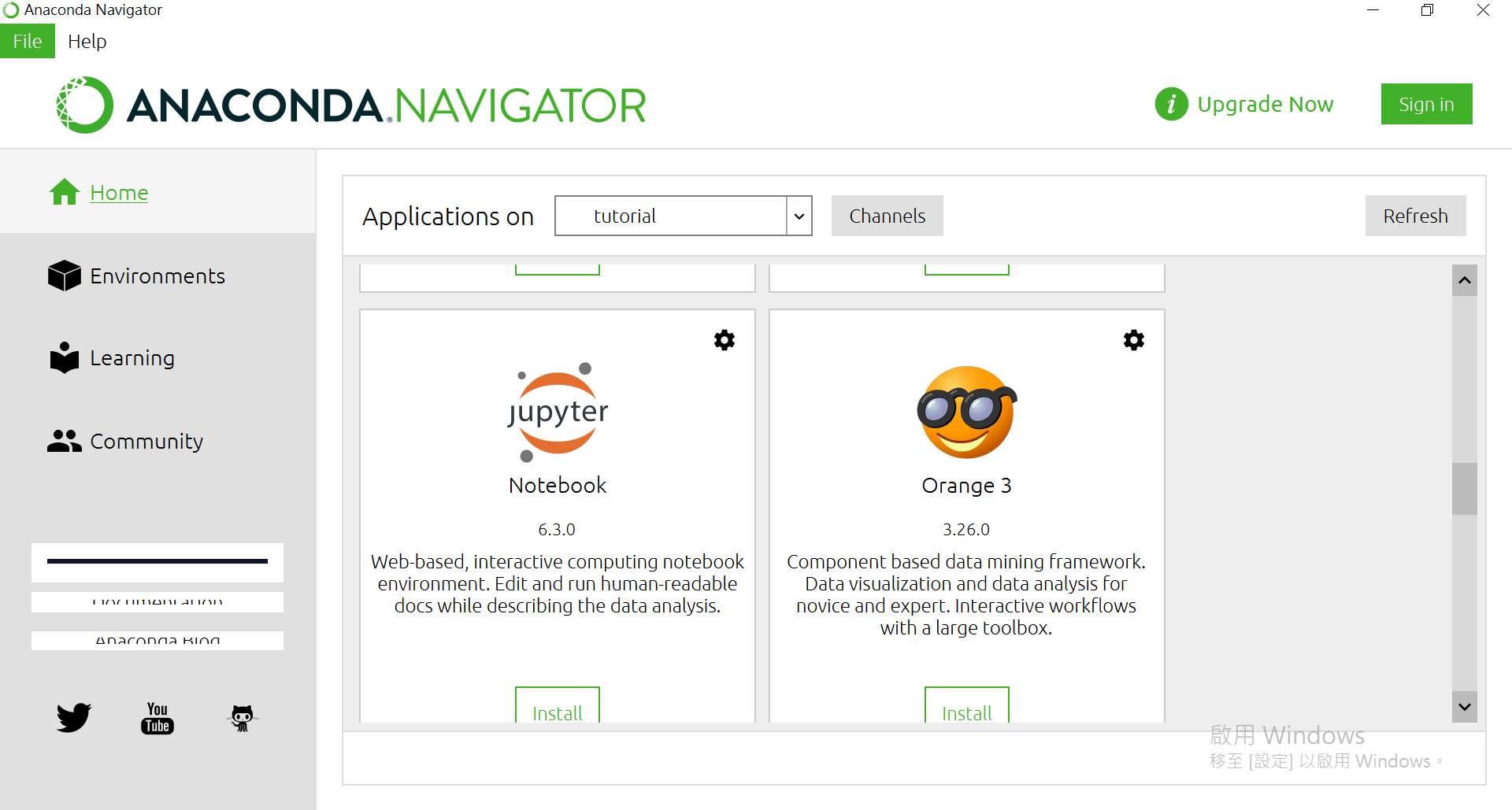
安裝完後可以打開。會從瀏覽器開在本地端的伺服器(其實也可以對外遠端使用Jupyter)
初始畫面: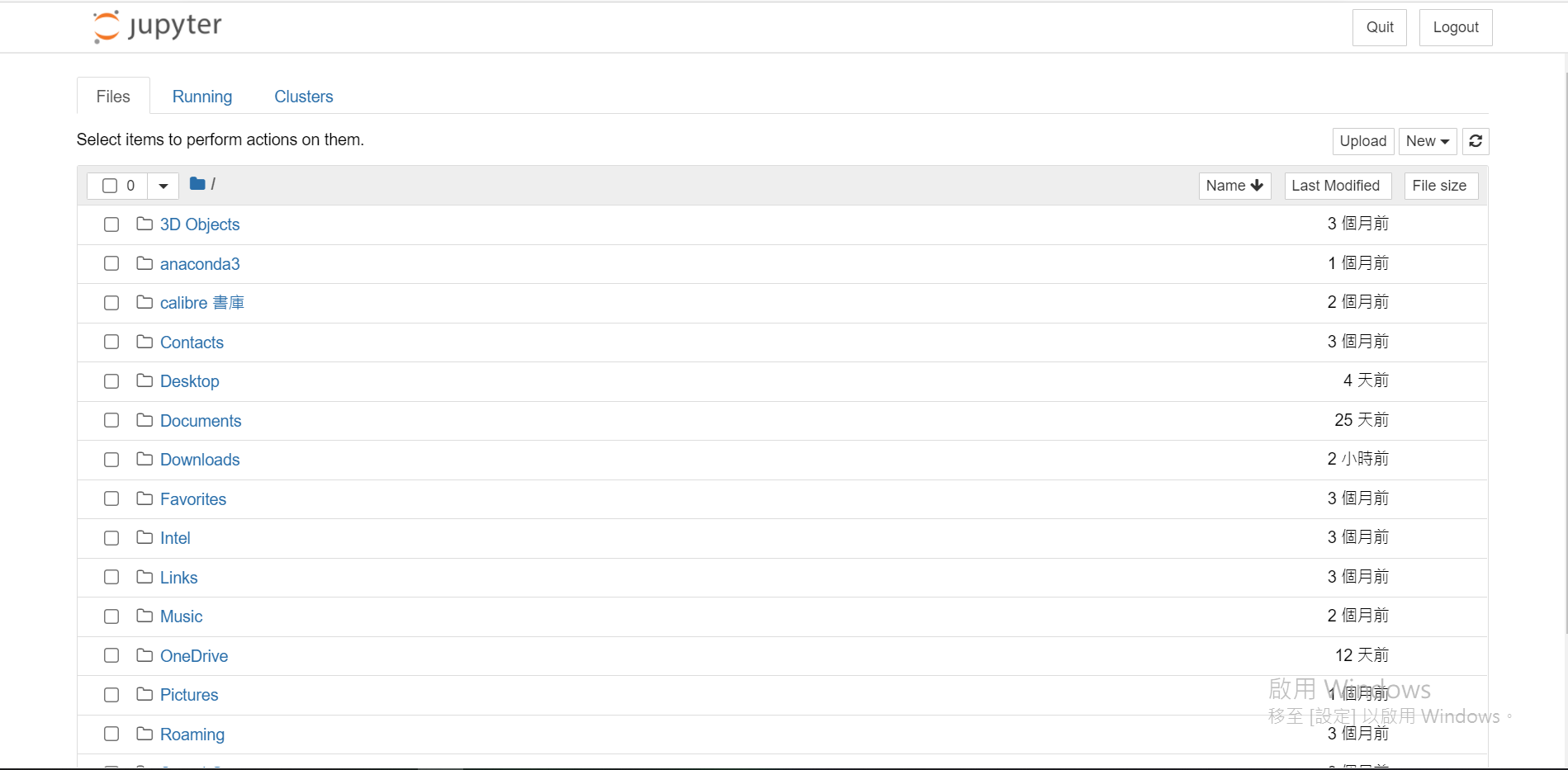
找到/建立一個你喜歡的資料夾(我是放在桌面拉,會叫做Desktop)
進到該資料夾後,右上方有個[New]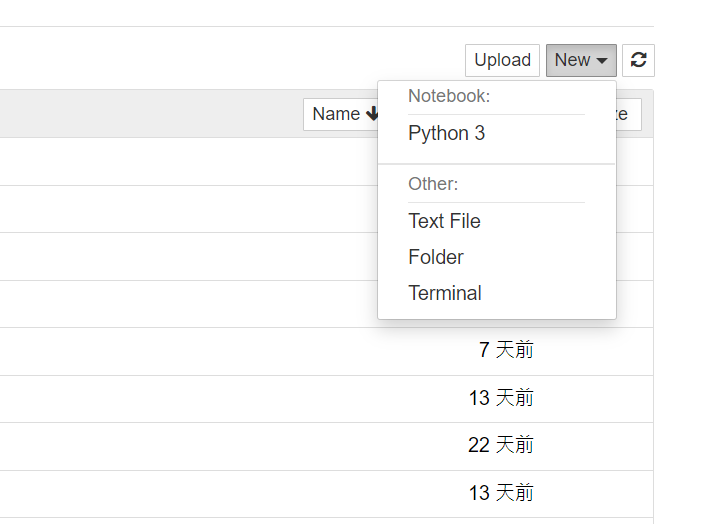
選擇Python就可以建立一個ipynb檔拉~
from sklearn import datasets, linear_model
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data
diabetes_y = diabetes.target
diabetes_X_train, diabetes_X_test, diabetes_y_train, diabetes_y_test = train_test_split(diabetes_X, diabetes_y, test_size=0.2)
model = linear_model.LinearRegression()
model.fit(diabetes_X_train, diabetes_y_train)
diabetes_y_pred = model.predict(diabetes_X_test)
print(f'R2 score: {r2_score(diabetes_y_test, diabetes_y_pred)}')
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LinearRegression
rng = np.random.RandomState(10)
x = 10 * rng.rand(30)
y = np.sin(x) + 0.1 * rng.randn(30)
poly_model = make_pipeline(PolynomialFeatures(7), LinearRegression())
poly_model.fit(x[:, np.newaxis], y)
xfit = np.linspace(0, 10, 100)
yfit = poly_model.predict(xfit[:, np.newaxis])
plt.scatter(x, y)
plt.plot(xfit, yfit)
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data
diabetes_y = diabetes.target
diabetes_X_train, diabetes_X_test, diabetes_y_train, diabetes_y_test = train_test_split(diabetes_X, diabetes_y, test_size=0.2)
model = linear_model.Ridge(alpha=1.0)
model.fit(diabetes_X_train, diabetes_y_train)
diabetes_y_pred = model.predict(diabetes_X_test)
print(f'R2 score: {r2_score(diabetes_y_test, diabetes_y_pred)}')
from sklearn import neighbors, datasets
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
iris = datasets.load_iris()
X = iris.data
y = iris.target
model = neighbors.KNeighborsClassifier()
model.fit(X, y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = neighbors.KNeighborsClassifier()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
from sklearn import preprocessing
iris = load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = DecisionTreeClassifier()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
from sklearn.ensemble import RandomForestClassifier
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.datasets import load_iris
iris = load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = RandomForestClassifier(max_depth=6, n_estimators=10)
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
from sklearn.datasets import load_breast_cancer
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
cancer = load_breast_cancer()
X = cancer.data
y = cancer.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = SVC(kernel='rbf')
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
from sklearn import datasets
from sklearn.naive_bayes import MultinomialNB
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
iris = datasets.load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = MultinomialNB()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
import numpy as np
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.utils import np_utils
#preparing data for Exclusive OR (XOR)
attributes = [
#x1, x2
[0, 0]
, [0, 1]
, [1, 0]
, [1, 1]
]
labels = [
[1, 0]
, [0, 1]
, [0, 1]
, [1, 0]
]
data = np.array(attributes, 'int64')
target = np.array(labels, 'int64')
model = Sequential()
model.add(Dense(units=3 , input_shape=(2,))) #num of features in input layer
model.add(Activation('relu')) #activation function from input layer to 1st hidden layer
model.add(Dense(units=3))
model.add(Activation('relu')) #activation function from 1st hidden layer to 2end hidden layer
model.add(Dense(units=2)) #num of classes in output layer
model.add(Activation('softmax')) #activation function from 2end hidden layer to output layer
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
score = model.fit(data, target, epochs=100)
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Convolution2D, MaxPooling2D
from keras.utils import np_utils
from keras.datasets import mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train = X_train.reshape(X_train.shape[0],28, 28, 1)
X_test = X_test.reshape(X_test.shape[0], 28, 28, 1)
Y_train = np_utils.to_categorical(y_train, 10)
Y_test = np_utils.to_categorical(y_test, 10)
model = Sequential()
model.add(Convolution2D(32, (3, 3), activation='relu', input_shape=(28,28,1)))
model.add(Convolution2D(32, (3, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(10, activation='softmax'))
model.compile(loss='categorical_crossentropy',optimizer='adam',metrics=['accuracy'])
model.fit(X_train, Y_train, batch_size=32, epochs=1, verbose=1)
score = model.evaluate(X_test, Y_test)
print('Test accuracy: {}'.format(score[1]))
==== 待更 ====
