除了 EM segmentatation challenge 比賽之外,還有參加了另外一個比賽 ISBI cell tracking challenge,應用了兩個資料集。
(1) PhC-U373
(2) DIC-HeLa
We also applied the u-net to a cell segmentation task in light microscopic images. This segmenation task is part of the ISBI cell tracking challenge 2014 and 2015 [10,13].
說明了還去參加了什麼樣的比賽。
The first data set “PhC-U373”2 contains Glioblastoma-astrocytoma U373 cells on a polyacrylimide substrate recorded by phase contrast microscopy (see Figure 4a,b and Supp. Material).
細胞的細節
It contains 35 partially annotated training images. Here we achieve an average IOU (“intersection over union”) of 92%, which is significantly better than the second best algorithm with 83% (see Table 2).
火力展示環節。
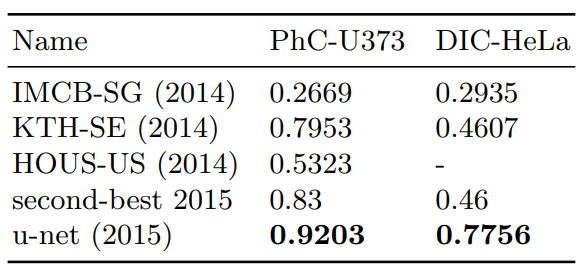
The second data set “DIC-HeLa”3 are HeLa cells on a flat glass recorded by differential interference contrast (DIC) microscopy (see Figure 3, Figure 4c,d and Supp. Material). It contains 20 partially annotated training images. Here we achieve an average IOU of 77.5% which is significantly better than the second best algorithm with 46%.
火力展示環節2。
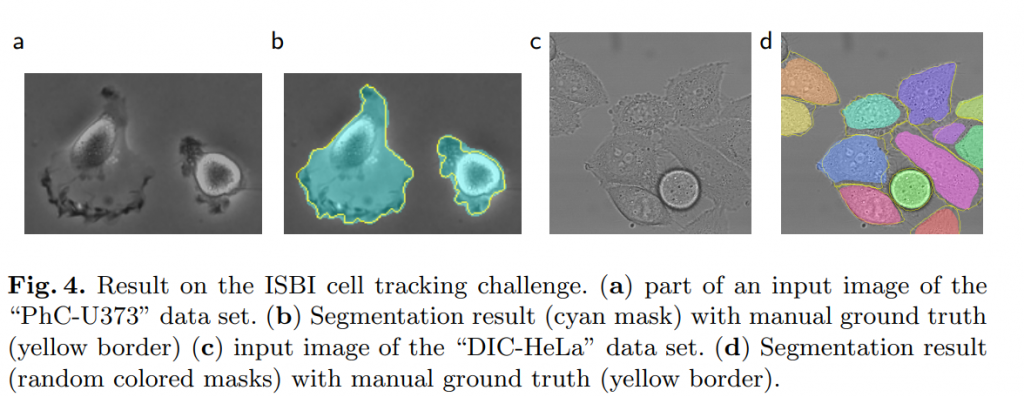
兩種資料級的樣子。
論文終於快讀完了,在實驗階段Unet沒有多說廢話,說明規則,直接說明自己有多強。
[0] U-net
