import pandas as pd
import numpy as np
import random
import socket
from enum import Enum
import matplotlib.pyplot as plt
HOST = '127.0.0.1'
PORT = 8888
s = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
s.connect((HOST, PORT))
class CrossoverType(Enum):
single = 1
double = 2
more = 3
class MutationType(Enum):
Inversion = 1
Insertion = 2
Displacement = 3
ReciprocalExchange = 4
class SelectionType(Enum):
Deterministic = 1
Stochastic= 2
class GeneticAlgorithm:
def __init__(self,pop_size,number_of_genes,selection_type,
crossover_type,crossover_rate,mutation_type,mutation_rate,
):
self.pop_size = pop_size
self.selection_type = selection_type
self.crossover_size = int(pop_size*crossover_rate)
if(self.crossover_size%2==1):
self.crossover_size -= 1;
self.mutation_size = int(pop_size*mutation_rate)
self.total_size = self.pop_size+self.mutation_size+self.crossover_size
self.number_of_genes = number_of_genes
self.crossover_type = crossover_type
self.crossover_rate = crossover_rate
self.mutation_type = mutation_type
self.mutation_rate = mutation_rate
def initialize(self):
self.selected_chromosomes = np.zeros((self.pop_size,self.number_of_genes))
self.indexs = np.arange(self.total_size) #如果np.arrange(5),輸出為array([0,1,2,3,4])
self.chromosomes = np.zeros((self.total_size,self.number_of_genes),dtype=int)
for i in range(self.pop_size):
rtv = [random.randint(0,1) for _ in range(self.number_of_genes)]
for j in range(self.number_of_genes):
self.chromosomes[i][j] = rtv[j]
self.fitness = np.zeros(self.total_size)
self.objective_values = np.zeros(self.total_size)
self.best_chromosome = np.zeros(self.number_of_genes,dtype=int)
self.best_fitness = 0
def evaluate_fitness(self):
for i,chromosome in enumerate(self.chromosomes[:self.pop_size]):
self.objective_values[i] = self.countbyflexsim(chromosome)
def update_best_solution(self):
best_index = np.argmin(self.objective_values)
if(self.best_fitness < self.objective_values[best_index]):
self.best_fitness = self.objective_values[best_index]
for i,gene in enumerate(self.chromosomes[best_index]):
self.best_chromosome[i] = gene
def countbyflexsim(self,scenario):
list = scenario.tolist()
scenario_str = str(list)[1:-1]
s.send(scenario_str.encode())
indata = s.recv(1024)
indata = indata.decode()
indata = indata.strip()
indata = indata.strip(',')
indata = float(indata)
return indata
def shuffle_index(self,length):
for i in range(length):
self.indexs[i] = i
np.random.shuffle(self.indexs[:length])
def perform_crossover_operation(self):
self.shuffle_index(self.pop_size)
child1_index = self.pop_size
child2_index = self.pop_size+1
count_of_crossover = int(self.crossover_size/2)
for i in range(count_of_crossover):
parent1_index = self.indexs[i]
parent2_index = self.indexs[i+1]
if(self.crossover_type == CrossoverType.single):
self.single_point_crossover(parent1_index,parent2_index,child1_index,child2_index)
self.objective_values[child1_index] = self.countbyflexsim(self.chromosomes[child1_index])
self.objective_values[child2_index] = self.countbyflexsim(self.chromosomes[child2_index])
elif(self.crossover_type == CrossoverType.double):
self.double_point_crossover(parent1_index,parent2_index,child1_index,child2_index)
self.objective_values[child1_index] = self.countbyflexsim(self.chromosomes[child1_index])
self.objective_values[child2_index] = self.countbyflexsim(self.chromosomes[child2_index])
else:
self.more_point_crossover(parent1_index,parent2_index,child1_index,child2_index)
self.objective_values[child1_index] = self.countbyflexsim(self.chromosomes[child1_index])
self.objective_values[child2_index] = self.countbyflexsim(self.chromosomes[child2_index])
child1_index +=2
child2_index +=2
def single_point_crossover(self,p1,p2,c1,c2):
self.chromosomes[c1] = self.chromosomes[p1]
self.chromosomes[c2] = self.chromosomes[p2]
cross_point = np.random.randint(1,self.number_of_genes)
self.chromosomes[c1][cross_point:] = self.chromosomes[p2][cross_point:]
self.chromosomes[c2][cross_point:] = self.chromosomes[p1][cross_point:]
def double_point_crossover(self,p1,p2,c1,c2):
self.chromosomes[c1] = self.chromosomes[p1]
self.chromosomes[c2] = self.chromosomes[p2]
index = []
ran = random.sample(range(0,self.number_of_genes),2)
for i in ran:
index.append(i)
index.sort()
self.chromosomes[c1][index[0]:index[1]] = self.chromosomes[p2][index[0]:index[1]]
self.chromosomes[c2][index[0]:index[1]] = self.chromosomes[p1][index[0]:index[1]]
def more_point_crossover(self,p1,p2,c1,c2):
self.chromosomes[c1] = self.chromosomes[p1]
self.chromosomes[c2] = self.chromosomes[p2]
index = []
ran = random.sample(range(0,self.number_of_genes),3)
for i in ran:
index.append(i)
index.sort()
self.chromosomes[c1][index[0]:index[1]] = self.chromosomes[p2][index[0]:index[1]]
self.chromosomes[c1][index[2]:self.number_of_genes] = self.chromosomes[p2][index[2]:self.number_of_genes]
self.chromosomes[c2][index[0]:index[1]] = self.chromosomes[p1][index[0]:index[1]]
self.chromosomes[c2][index[2]:self.number_of_genes] = self.chromosomes[p1][index[2]:self.number_of_genes]
def do_roulette_wheel_selection(self,fitness_list):
sum_fitness = sum(fitness_list)
transition_probability = [fitness/sum_fitness for fitness in fitness_list]
rand = random.random() #輸出為0-1範圍的實數
sum_prob = 0
for i,prob in enumerate(transition_probability):
sum_prob += prob
if(sum_prob>=rand): #當第一個位置的機率條件符合 回傳遞第一個位置
return i
def perform_selection(self):
if self.selection_type == SelectionType.Deterministic:
index = np.argsort(self.objective_values)[::1]
elif self.selection_type == SelectionType.Stochastic:
index = [self.do_roulette_wheel_selection(self.objective_values) for i in range(self.pop_size)]
else:
index = self.shuffle_index(self.total_size)
for i in range(self.pop_size):
for j in range(self.number_of_genes):
self.selected_chromosomes[i][j] = self.chromosomes[index[i]][j]
for i in range(self.pop_size):
for j in range(self.number_of_genes):
self.chromosomes[i][j] = self.selected_chromosomes[i][j]
def perform_mutation_operation(self):
self.shuffle_index(self.pop_size+self.crossover_size)
child1_index = self.pop_size+self.crossover_size
for i in range(self.mutation_size):
if(self.mutation_type==MutationType.Inversion):
parent1_index = self.indexs[i]
self.shuffle_mutation(parent1_index,child1_index)
self.objective_values[child1_index] = self.countbyflexsim(self.chromosomes[child1_index])
child1_index += 1
def shuffle_mutation(self,p1,c1):
for i in range(self.number_of_genes):
self.chromosomes[c1][i] = self.chromosomes[p1][i]
ran = random.sample(range(0,self.number_of_genes),12)
for i in ran: #隨機選擇12個基因進行突變
if(self.chromosomes[c1][i]==0):
self.chromosomes[c1][i] = 1
else:
self.chromosomes[c1][i] = 0
pop_size = 2
selection_type = SelectionType.Stochastic
crossover_type = CrossoverType.more #single double more
crossover_rate = 1
mutation_type = MutationType.Inversion
mutation_rate = 0.5
number_of_genes =60
solver = GeneticAlgorithm(pop_size,number_of_genes,selection_type,
crossover_type,crossover_rate,
mutation_type,mutation_rate,
)
solver.initialize()
solver.evaluate_fitness()
for i in range(100):
solver.perform_crossover_operation()
solver.perform_mutation_operation()
solver.update_best_solution()
solver.perform_selection()
if(i %10 ==0):
print(F"iteration {i} :")
print(f"{solver.best_chromosome}: {solver.countbyflexsim(solver.best_chromosome)}")
outdata = "exit"
s.send(outdata.encode())
以上是我基因演算法的程式碼
我想請問如何調整程式碼能夠畫出類似以下的圖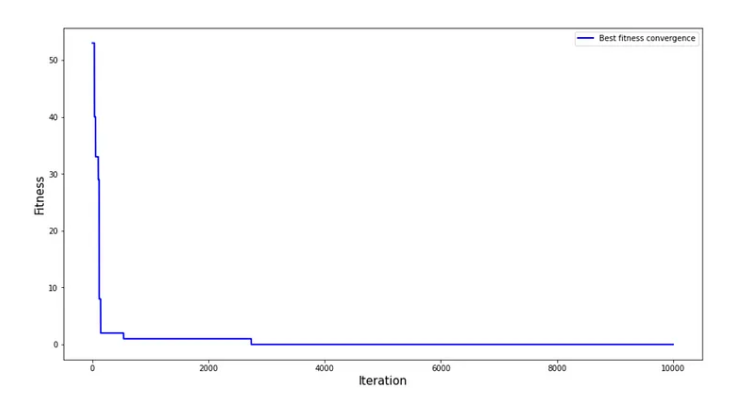
X軸為iteration
Y軸為當次iteration最佳適應值也就是best_fitness
